Design CRISPR/Cas9 sgRNA
Supercomputer-based web server
Whole genome screening of on-target and off-target sites
Machine learning based evaluation system
Genome wide visualization
Get started!
Supercomputer-based web server
Whole genome screening of on-target and off-target sites
Machine learning based evaluation system
Genome wide visualization

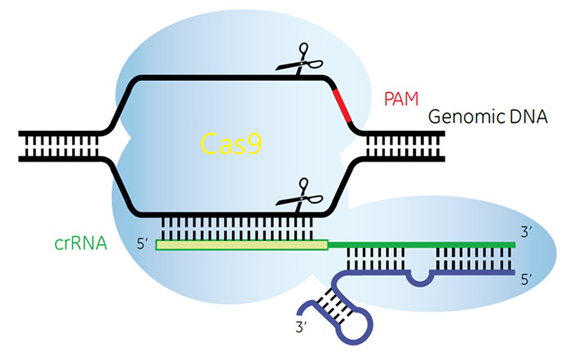
CRISPR is a system widely used for mammalian genome editing. However, Most of the existing design tools are mainly focused on on-target sites evaluation, seldom of them can predict off-target effect with sufficient efficiency and accuracy. To address this issue, we developed CRISPR Designer that is dedicated to aid the design of CRISPR/Cas9 sgRNA for any gene of interest. For the support of a large user volume, the web server is exported to Tianhe-2 supercomputer and achieved significant performance improvement.
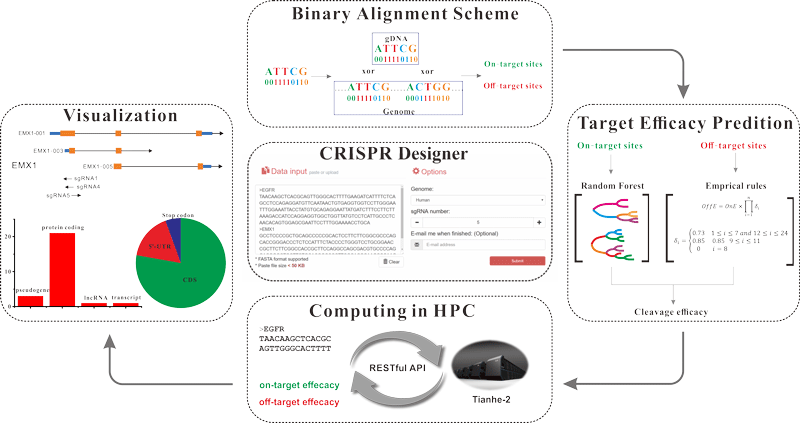
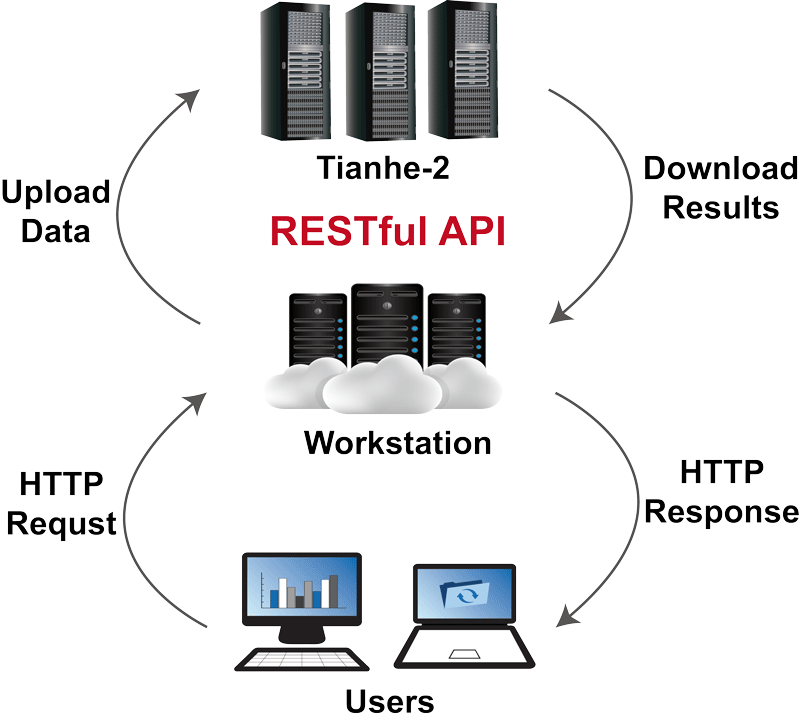
To support a large user volume, we have developed a parallel version of the CRISPR Designer and exported it to the Tianhe-2 supercomputer. Taking advantage of Tianhe-2’s massive computing power, CRISPR Designer can support over 200 simultaneous users and speed up the identification of potential target sites by conducting the computational task in multi-core CPU.
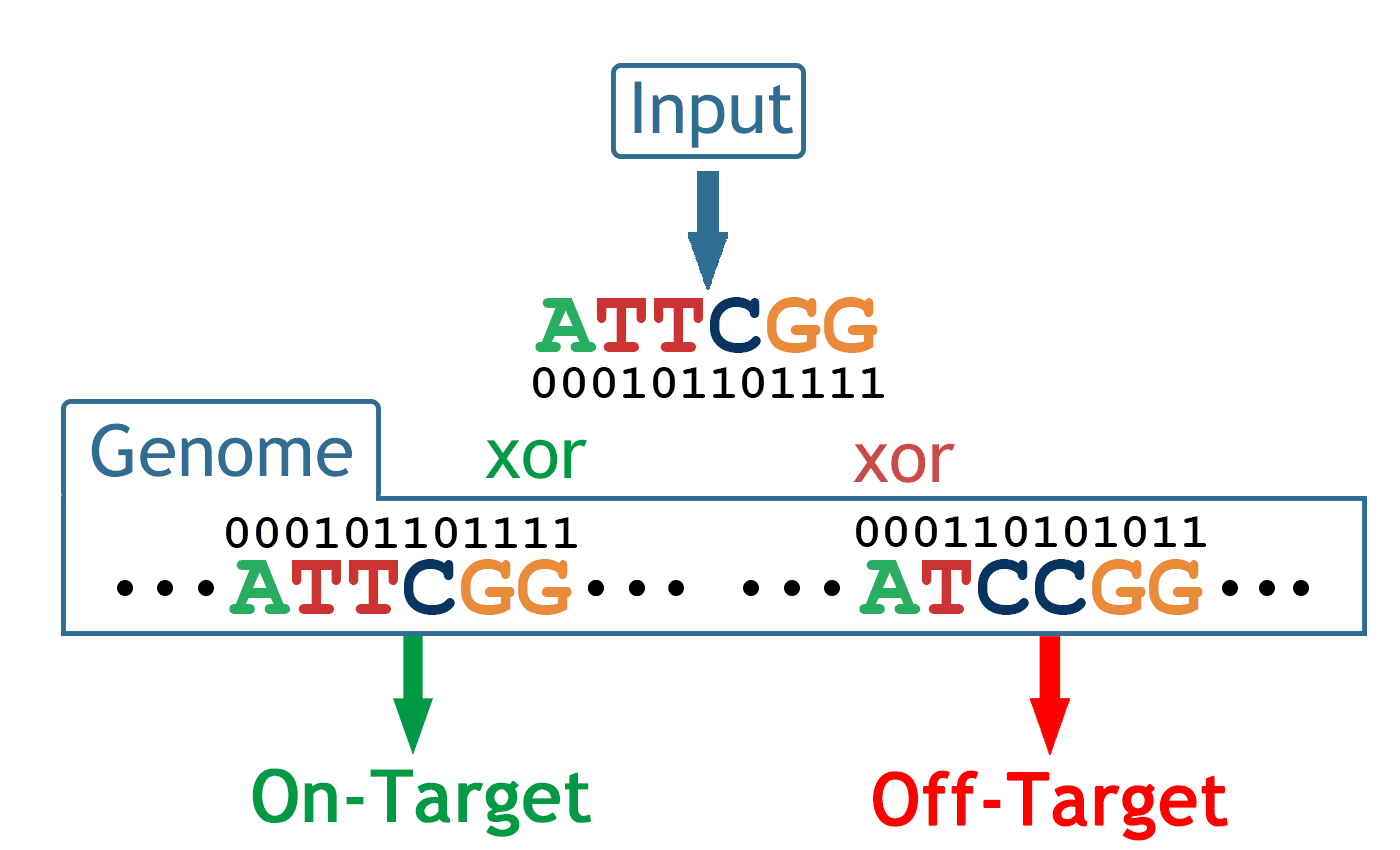
By implementing a binary alignment scheme, CRISPR Designer is able to efficiently screen the whole genome and then identify potential on-target and off-target sites in just a few minutes.
To maximize on-target efficacy and minimize off-target effect, we applied the Random Forest algorithm for quantitatively assessing both the on-target and off-target activity. The evaluation on additional test sets have proven that the on-target and off-target models can reach an accuracy of 68.7% and 88.0%, respectively. Based on the predicted activities, a combination score is then calculated, according to which the best optimized sgRNA designs are reported.
 Baker, Paul T. et al. Phys.Rev 2015
Baker, Paul T. et al. Phys.Rev 2015
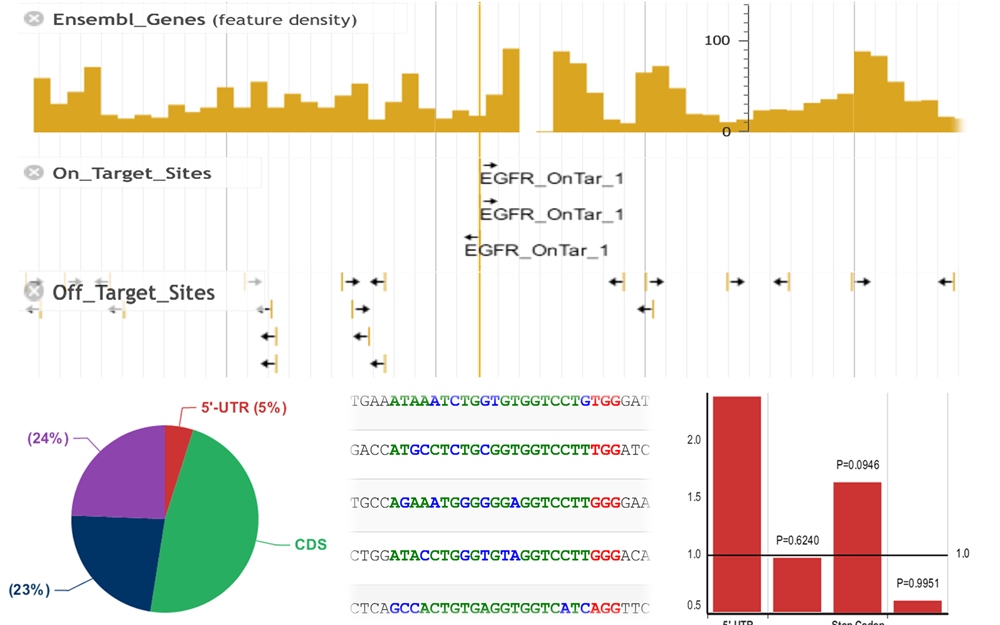
To facilitate the visualization of potential target sites, we integrated a genome browser and several statistical diagrams in the web server to display the distribution of target sites in genome according to their precise coordinates.